There is a new post in Pymol user mailing list showing an approach to draw the “Goodsell-like” molecule. The most famous Goodsell molecular images are the “Molecule of the month” at PDB-101. Dr. Goodsell also provides online version of images used in his book: “The Machinery of Life“.
That detail of Goodsell-like presentation in pymol is available at PyMOLWiki. It mainly uses GLSL rendering technique. I practiced several times and found out that my MacPyMOL version (2 years old) is not able to run such rendering (OpenGL only..). Therefore I used macports to install PyMOL 1.5 (run under X11).
There may be a wrong description at PyMOLWiki — I rename the “sphere.fs” to “sphere.fs.bak” and copy and save the author’s code (at PyMOLWiki) as “sphere.fs” (not shaders.fs).
Here is another 2 tries I have done using PDB files 2Y1M and 2WCC for top 2 figures and the bottom DNA figure, respectively.
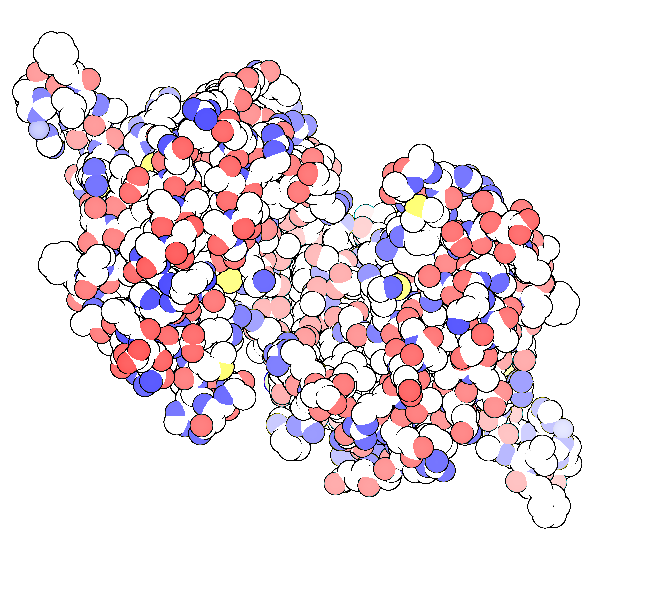



Thank you. I asked you about it a long time back. I like this kind of representation so much. Actually I found another way to do it. You can get the same output in PMV (of The Scripps Research Institute) with just a single click. The cartoon representation of CPK model there produces similar image. It’s always better to know more than one way to do a thing. Thanks 🙂
Comment by uttam2707 — June 28, 2012 @ 1:43 am |
Yes, I did see your question long time ago and I tried using PyMOL to draw such Goodsell-like image. Unfortunately I don’t find a way at that time. PMV is a good molecular viewer–recently found it,too.
Comment by kpwu — June 28, 2012 @ 11:21 am |
[…] https://kpwu.wordpress.com/2012/06/28/pymol-draw-goodsell-like-view-using-glsl/ […]
Pingback by David Goodsell like images | Getting to know Structural Bioinformatics — April 30, 2013 @ 5:47 pm |