Here is an example using “grid mode” provided in PyMOL to show the charged states of a protein (PDB entry: 1Z66) at pH 4 (left), 7 (center) and 11 (right). PyMOL script is attached after the figure.
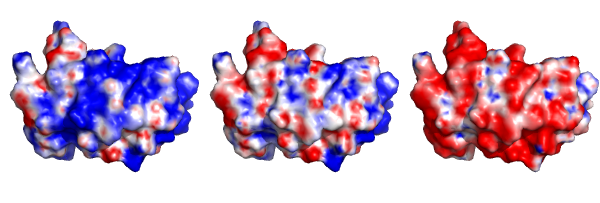
First, the potential maps at three pH states have to be generated. I used APBS web server combined with PROPKA to get calculated pH-dependent potentials (.dx files).
Then I load 1z66.pdb and load all 3 .dx files. Draw first protein with one of the potentials. After that, turn on the grid mode and then duplicate pdb object twice.
Last, assign the potentials to duplicated objects. The electrostatic surface views at 3 pH states can be shown easily!
I really like the grid mode in PyMOL! No need to use any image editor to assemble 3 potential views. No worry of sizes of molecules if generated separately.
Tips of “grid mode” can be found in my earlier post: [PyMOL] grid mode.
Steps to “Draw the electrostatic surface” can be checked at my earlier post, too.
=======pymol log.pml =================
load 1z66.pdb
load 1z66-pH4.dx, emap_pH4
load 1z66-pH7.dx, emap_pH7
load 1z66-pH11.dx, emap_pH11
set grid_mode, on
cmd.show_as(“surface” ,”1z66″)
ramp_new elvl1, emap_pH4, [-5,0,5]
set surface_color, elvl1, 1z66,
cmd.disable(‘elvl1’)
cmd.create(None,”1z66″)
ramp_new elvl2, emap_pH7, [-5,0,5]
set surface_color, elvl2, obj01,
cmd.create(None,”1z66″)
ramp_new elvl3, emap_pH11, [-5,0,5]
set surface_color, elvl3, obj02,
cmd.disable(‘elvl3’)
cmd.disable(‘elvl2’)
bg_color white
ray 600
png 1z66.png















