Follow up previous post, here shows several modes available in PyMOL to display the ribbon/chain of DNA/RNA.
Example molecule: B-form DNA PDB code: 2L8Q
Running version, PyMOL 1.3 (x11 version on MacOS 10.6)
The previous version use “oval” to show the ribbon of DNA. Here are some other ways to show the ribbon:
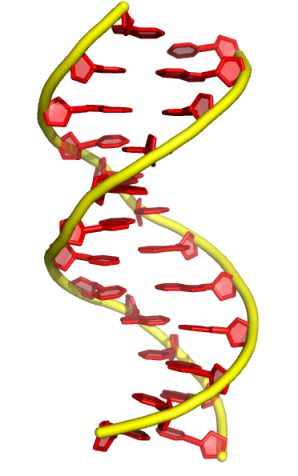
1. default ribbon shown by cartoon (“tube” mode)
default radius of tube mode
set cartoon_tube_radius, 1
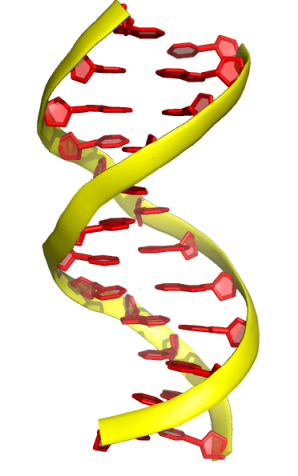
2. The “oval” mode
cartoon oval
set cartoon_oval_width, 0.3
set cartoon_oval_length, 1.2
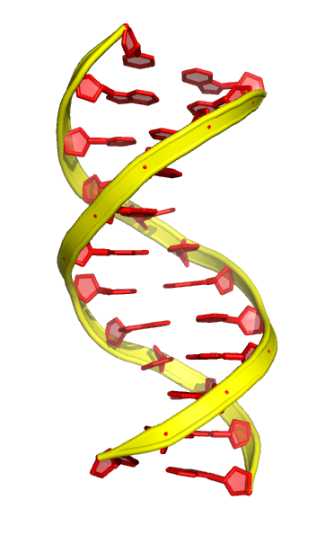
3. The “rect” mode:
cartoon rect
set cartoon rect_width, 0.5
set cartoon rect_length, 2
combined with addition command (molecule rotated):
cartoon arrow
4. The “dumbbell” mode:
cartoon dumbbell,
set cartoon_dumbbell_width, 0.3
set cartoon_dumbbell_length, 1
set cartoon_dumbbell_radius, 0.35
In most protein structures shown in papers, the helices and sheets are always colored in two colors. We can use “cartoon_highlight_color” to create such effect on DNA,too. Here is the example:
set cartoon_highlight_color, navy
set cartoon_nucleic_acid_color, palecyan
2012 May 24, new post about different colors for the rings.