In PyMOL Wiki, there is a “contact map visualizer” plugin providing users to inspect the protein intramolecular contacts. However, this plugin requires “PyGame”, “Tkinter”, “PIL (python image library)” and most importantly, Gromacs!
I used Fink (Mac OSX 10.6.8) to successfully install Gromacs 4.1 but Fink reported problems while installing pygame. So I can generate xpm file of target protein structure by Gromacs but not inspect the contact map in PyMOL.
In contrast, I found another contact map visualizer called “CMView – Protein contact map visualization and analysis“. This program is written in Java and uses PyMOL as the molecular viewer.
Installation note (work platform: Mac OSX 10.6.8 with X11)
1. Download the CMView file, unzip it and move the folder to proper path)
2.Edit the “cmview.cfg” file and correct the path of pymol execute. Optional programs such as DSSP and DALI can be used if installed.
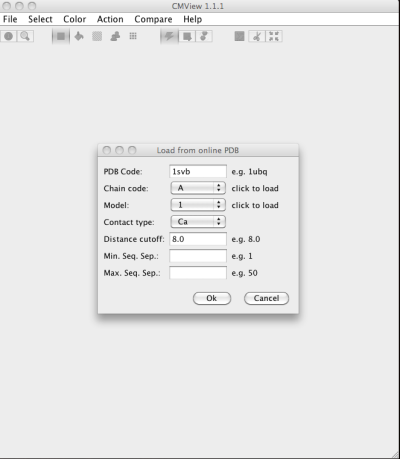
3. In terminal, call “cmview.sh” and load the PDB file from “CMView” and enjoy visualizing the molecules.
While the PDB is loaded, the CMView (run in Java) will show you the contact map. You can use mouse to crop the region of interest and instantly see the contacts in the PDB file.


Can you export the contact map plot with axes (name “Number of residue” and enumeration)?
Comment by murpholinox peligro (@murpholinox) — June 26, 2012 @ 10:57 am |
I guess it’s not provided by the CMView. Hope the author will provide such information while generating output in the newer version.
Comment by kpwu — June 27, 2012 @ 1:03 am |
Very Excellent
Comment by suvarnavanik — April 5, 2013 @ 2:05 am |